CRISPR - Changing the gene editing landscape
By: Dong Liu
In the last few years, the scientific community has seen rapid development of a new and ‘game-changing’ technology called CRISPR/Cas9. But what is CRISPR? How is it is being used by stem cell researchers? And why has it sparked debate? In this post, Dong Liu (PhD student at MRC Centre for Regenerative Medicine) describes a technology that is altering the pace and possibilities of stem cell research.
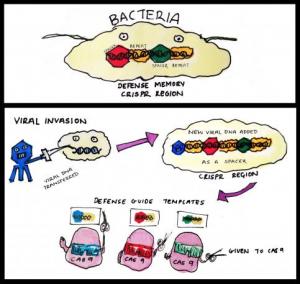
A Bug’s Life – CRISPR technology was discovered in bacteria
The term CRISPR (clustered regularly interspaced short palindromic repeats) was coined in 2002 by Dutch scientists working on bacteria genomes. It describes regions of the genome that contain multiple copies of DNA repeats separated by unique ‘spacers’. Though its discoverers had few clues at the time about what these curious sequences do, it has become apparent that these DNA elements represent a form of bacterial immunity against pathogens like viruses.
Much like our own immune system, CRISPR regions store the memory of previous infections and help producing a stronger response when invaded again. Viral DNA seized by the bacteria upon initial infection is incorporated into the CRISPR regions to become the spacers. These hybrid chunks of native (repeats) and foreign (spacers) sequences are then expressed in the form of short RNA molecules. These aptly named ‘guide RNAs’ can form a larger ‘seek and destroy’ structure when conjoined with an enzyme called Cas9. Cas9 is a nuclease, a type of molecular scissors that cut DNA. In the event of a second infection by the same virus, the guide RNA recognises the specific sequence and this results in Cas9 chopping up the viral DNA. This elegant mechanism therefore prevents repeated viral invasions.
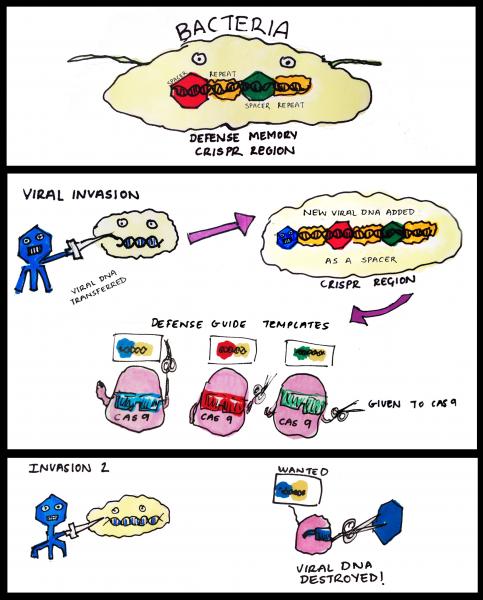
How CRISPR-Cas9 works in bacteria. CRISPR region in bacteria genome stores information from previous viral infections. Guide RNA made using these information allows Cas9 to 'see' viruses that had invaded before. Once identified, the viral DNA is rapidly destroyed by Cas9.
Our knowledge of how CRISPR works in bacteria is still incomplete, but the take home message is that CRISPR-Cas9 excels at cutting precise DNA sequences. This efficiency, combined with easier preparation of the molecular tools needed, is what makes this system so popular for biologists studying gene function.
A step-change in the speed of gene editing in the lab
To understand how genes influence cellular behaviour during development, normal physiology and disease, scientists have often adopted a ‘reverse genetics’ approach – they infer the functions of a particular gene from the effects of getting rid of it. In other words studying genes ‘knocked out’ in cells or model organisms. The process of generating specific gene knockouts is known as gene targeting or gene editing.
Traditional gene targeting was cumbersome and inefficient. It could take a skilled molecular biologist up to a few years to generate the tools needed and then produce a knockout animal.
The prospect improved massively when scientists discovered a window of opportunity to edit genes – when the two strands of DNA break. DNA breaks are harmful to the cell, so they have to be rectified by the cell’s DNA damage repair molecules.
Using a ‘copy-and-paste’ mechanism, DNA sequences similar to those around the DNA break site can act as templates in repair. This reaction makes sure the template is precisely copied into the repaired sequence.
Another ‘quick-and-dirty’ way to repair is by simply joining the loose ends – an error-prone process which often leads to the disruption of the gene. Though lacking in subtlety, this mechanism is highly efficient when it comes to knocking out the genes of interest.
Which type of repair the cell undergoes is not entirely up to us, but the discovery of these natural mechanisms has allowed us to focus our gene targeting efforts on the first few steps of the process – introducing DNA breaks at specific places in the genome.
As such, the race to find better ways to edit genes became a matter of tackling how to make accurate DNA cuts. Artificial nucleases such as ZFNs and TALENs led the way in proof of principle, but they needed significant expertise to use – something CRISPR/Cas9 overcomes.
CRISPR is a valuable tool for researchers
Genetic modifications made easy
In a compelling demonstration of the efficiency of CRISPR, scientists at University of Tsukuba attempted to knockout a gene involved in the production of melanin, the pigment giving colour to our skin and hair. Following the injection of Cas9 and guide RNA into the embryos, 28 out of the 60 mice born were completely albino, suggesting that both copies of the gene had been disrupted. Remarkably, the timescale from CRISPR design to embryo injection in such a project is in the region of only a few weeks.
The high efficiency of CRISPR also allows scientists to target multiple genes in one step, and genome-wide CRISPR tools have been generated to facilitate screens potentially studying thousands of genes.

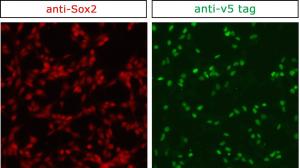
More convenient protein recognition
Proteins carry out most of the cell’s functions, so understanding where they are located and which molecules they interact with is no small matter. Antibodies that recognise the protein of interest are a standard part of the molecular and cellular biologist’s toolbox. However, the quality of commercial antibodies can be quite variable, and poor quality antibodies make it difficult to purify and visualise the target proteins. Using CRISPR, short biochemical tags can be introduced into a protein with ease by editing the gene encoding it. It is then possible to directly study the protein with tried and tested antibodies already generated against the tags, bypassing the often tedious routine of producing, testing and optimising specific antibodies.
Promising system for disease modelling
The ability to precisely edit genes is also helpful for understanding disease. It allows us to ask – in the lab - whether a suspect gene is directly involved in the progression of the disease, and work out if correcting a mutation associated with the disease could alleviate the symptoms.
For example, in 2014 US scientists effectively cured a mouse model of a liver disease by altering just one nucleotide, the smallest unit of DNA, of a specific faulty gene. Despite only restoring the function of 1 in 250 cells, the modified cells went on to expand and reconstitute the diseased liver.
CRISPR has formed a formidable partnership with another technology, induced pluripotent stem cells (iPS cells). These stem cells can be generated from people with a particular condition, and then used to produce the specialised cell types in which the disease genes have their effects (e.g. beta globin in sickle cell anaemia). Researchers can then investigate the influence of the disease gene on cell function. Sometime the cells can be directly used in chemical screens, searching for new small molecules - potential drugs - that could prevent the disease (more readings: see end of the piece).
Controversies surrounding editing human embryos
So if CRISPR technology is so easy to apply, doesn’t that raise some big questions? For example, could CRISPR be used to cure hereditary disorders via human germline editing - altering DNA in human sperm, egg or embryos.
In comparison with adult tissue, penetrating cells in early embryos to deliver CRISPR/Cas9 is easier to achieve. And rather than introducing into a range of tissues all different in their composition, a single protocol could be standardised for treating many genetic disorders.
Nonetheless, critics have already pointed out difficult questions facing human germline editing, both technically – and maybe more importantly regarding ethics and safety. Editing the human germline has always been viewed as a red line that should not be crossed by most researchers. This has reawakened the debate over possible creation of ‘designer babies’ whose genomes are tinkered with not to reduce disease risks, but for specific physical traits deemed superior.
In fact, earlier in 2015 when researchers at Sun Yat-sen University used CRISPR to edit the HBB gene in non-viable human embryos, a world first, the rate of accurate modification was a disappointing 14.3%. Most embryos used in the study either did not survive, displayed no Cas9 cuts, or repaired the DNA break with the error-prone end joining pathway. The authors also found substantial undesirable off-target mutations. This study has brought forth intense debate.
Indeed, some scientists have argued that genetic screening in pre-implantation embryos is a more favourable alternative to CRISPR gene editing in this case, providing good results with few complications. Arguments like this raise the question: do the benefits of human embryo editing really outweigh the risks?
CRISPR is powerful and promising and will no doubt have a part to play in the future of medicine. Regulatory and ethical considerations are just beginning to catch up with science’s advances. As written in a recent article about using CRISPR to modify T cell genes:
“There’s actually well-trodden ground putting modified T cells into patients. There are companies out there already doing it and figuring out the safety profile, so there’s increasing clinical infrastructure that we could potentially piggyback on as we work out more details of genome editing,” Marson said. “I think CRISPR-edited T cells will eventually go into patients, and it would be wrong not to think about the steps we need to take to get there safely and effectively.” (Alexander Marson, University of California, San Francisco)
Any decision we make will have far-reaching impacts and must not be taken lightly. CRISPR may well be the difference between suffering and well-being, but to ensure a smooth path from benchtop to bedside, we must not stop exploring the benefits, flaws and wider implications of this new technology. There is plenty of work to do yet.
Further information
- The ethics of changing genes in the embryo - commentary by Sarah Chan
- Statement by International Consortium on Stem Cells, Ethics and Law - The Hinxton Group on gene editing of embryos
- A perspective on the statement by the Hinxton Group
Acknowledgements
This article was written by Dong Liu, edited by Jan Barfoot and Kate Doherty, and reviewed by Steve Pollard, MRC Centre for Regenerative Medicine. The illustration of CRISPR was produced by Vanessa de Mello, University of Aberdeen, the picture of the albino pups was used by kind permission from Dr Seiya Mizuno, University of Tsukuba. The picture of the protein tagging by CRISPR was provided by Raul Bressan, University of Edinburgh.